Satyasaran Changdar

I am Satyasaran Changdar, Postdoc, working in Food Process Modelling using Scientific Machine Learning in project ‘SmartClean’, Ingredient and Dairy Technology, Department of Food Science [University of Copenhagen] (https://www.ku.dk/english/) under the Supervisons of Prof.Serafim Bakalis and collaborted with Arla foods.
Previously, I worked [July 2021-Feb 2024] as a postdoc in Applied Machine learning in Plant physiology under the supervision Dr. Erik Bjørnager Dam, Professor, Department of Computer Science, University of Copenhagen and Dr. Kristian Thorup-Kristensen Professor, Department of Plant and Environmental Sciences, University of Copenhagen, Denmark and completed my Ph.D. in applied mathematics in 2019 at University of Calcutta under the supervison of Dr. Soumen De and Dr. Samiran Ghosh, master of technology in computer applications from Indian Institute of Technology, Delhi, India in 2008 and masters in mathematics at Indian Institute of Technology, Bombay, India in 2005. Before joining Postdoc, I was working as an associate professor in Institute of engineering and Management, Kolkata, India.
I specialize in Machine Learning Deep Learning, Physics-informed Machine Learning, and Data-driven Scientific Computing. I had been actively involved in the development of machine learning methods that extract patterns from multimodal agriculture data in crop science from RadiMax in RadiBooster project. I have been working also on fine-tuning large language models (LLMs) with retrieval-augmented generation (RAG) in generative AI.
I have been collaborating with my PhD supervisor Dr. Soumen De, Department of Applied Mathematics, University of Calcutta and have supervised Bivas Bhaumik (Currently Assistant Prifessor at Department of Mathematics, NIT Rourkela, during his PhD. Currently I am supervising Susmita Saha and Soumini Dolui for their PhD at University of Calcutta, India. For more details: Download CV
Current Research Projects
Project 1: RadiMax: Machine Learning for understanding plant root function
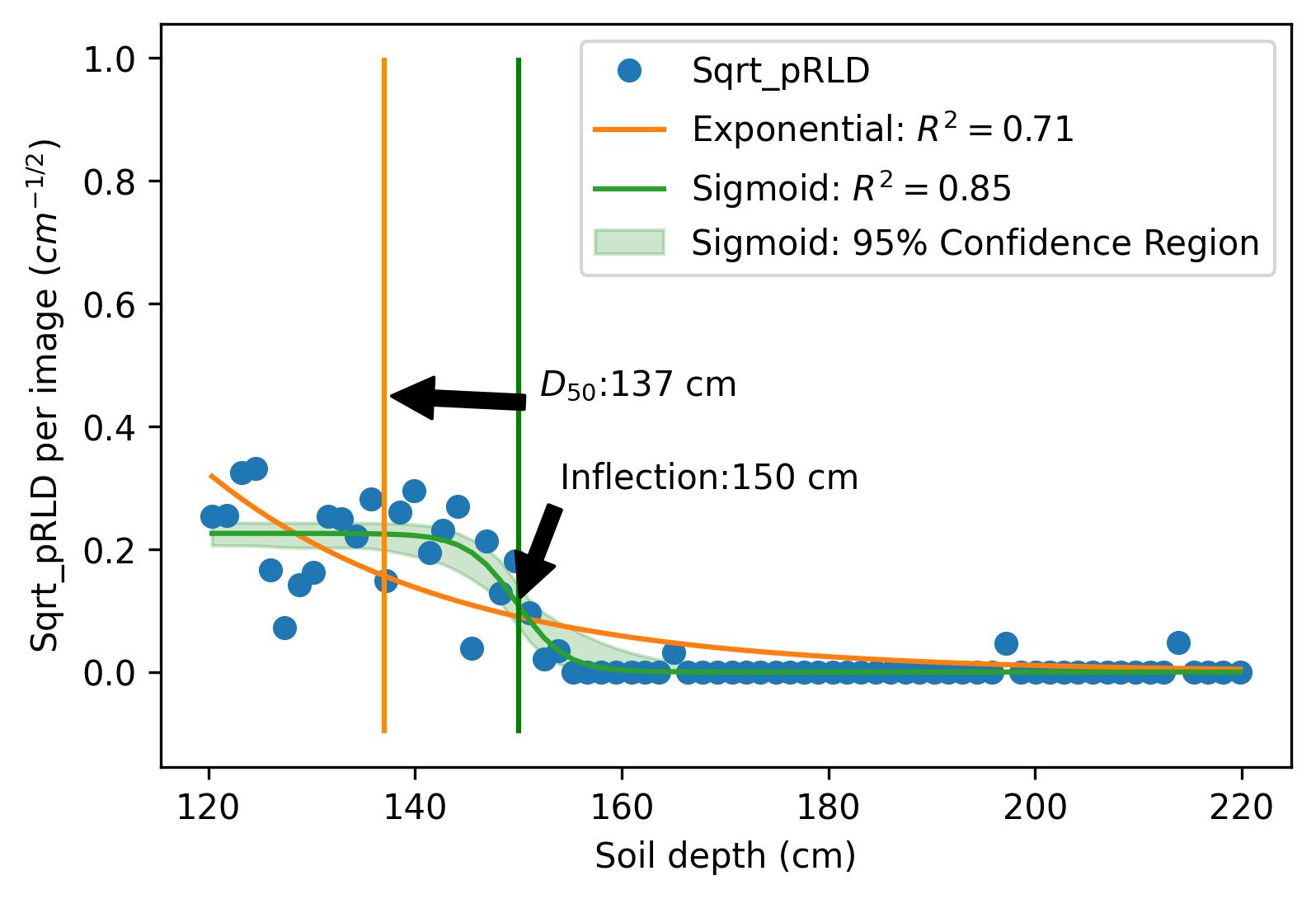
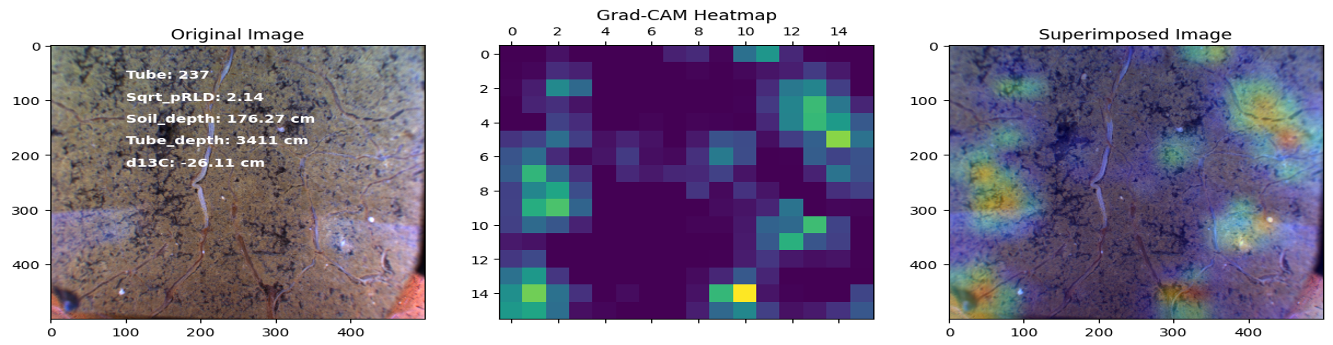
This project investigates the relationship between root distribution and resource uptake in crops using machine learning techniques. The study utilizes the RadiMax semi-field root-screening facility to phenotype winter wheat genotypes for root growth. Square root of planar root length [CNN, Deep learning was used to extract the root length from sub-soil images] density (Sqrt_pRLD) measurements are collected at different soil depths, and their correlation with deep soil nitrogen uptake and drought resilience potential is explored using machine learning models. The results demonstrate the importance of deep rooting for water and nitrogen uptake in crops.
Results
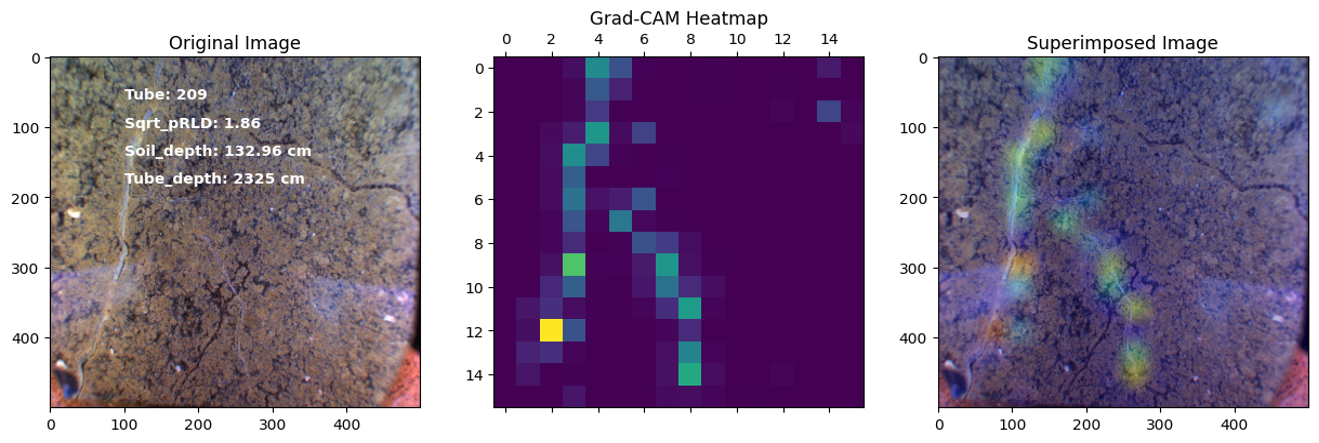
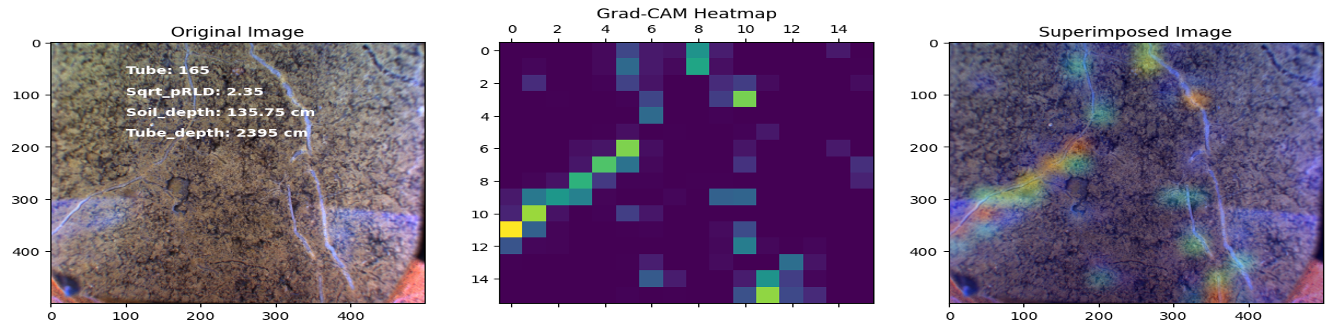
Project 2: RadiMax: Deep Learning for sub-soil Image analysis to investigate deep root function
In this project, our objective was to employ deep learning techniques to analyze sub-soil images and investigate the function of deep roots in plant physiology. Specifically, we utilized transfer learning with the ResNet50 architecture to develop regression and classification models for root length estimation and root function investigation.
Results
Here are some images showcasing the results:
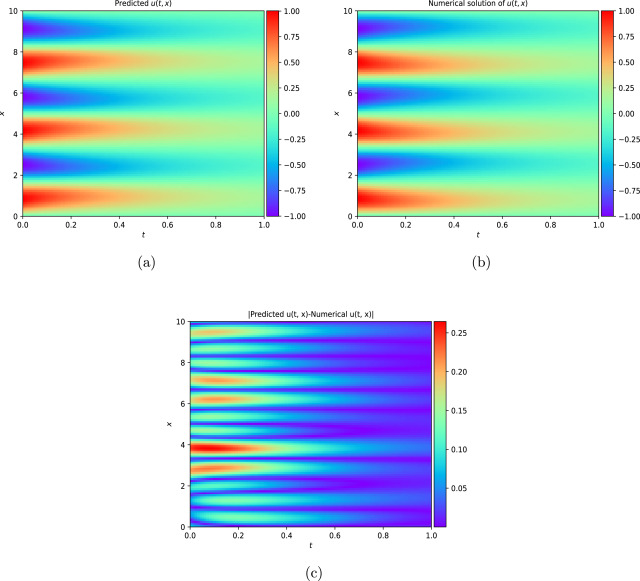
Project 3: Solving Non-linear partial differential equations in Blood flow modelling using Physics-informed Neural network
Paper link
Code Link
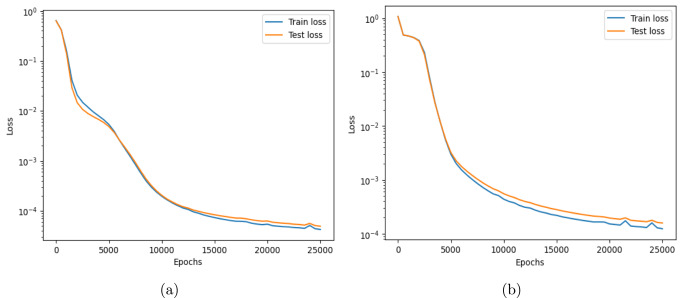
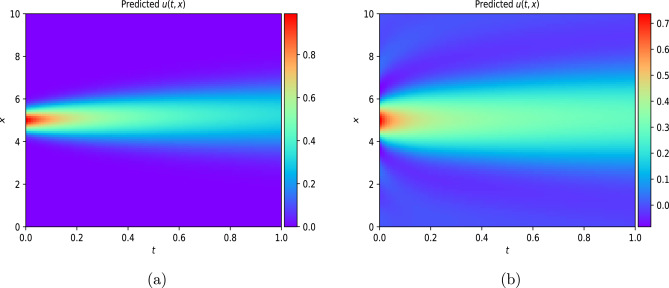
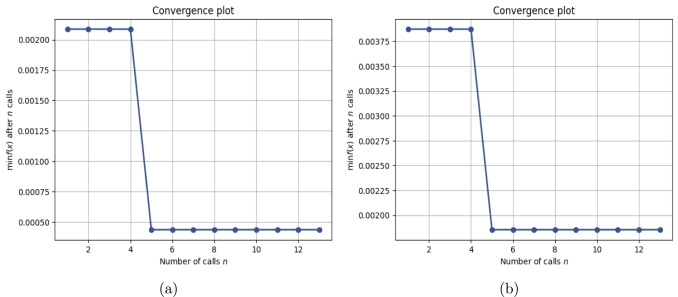
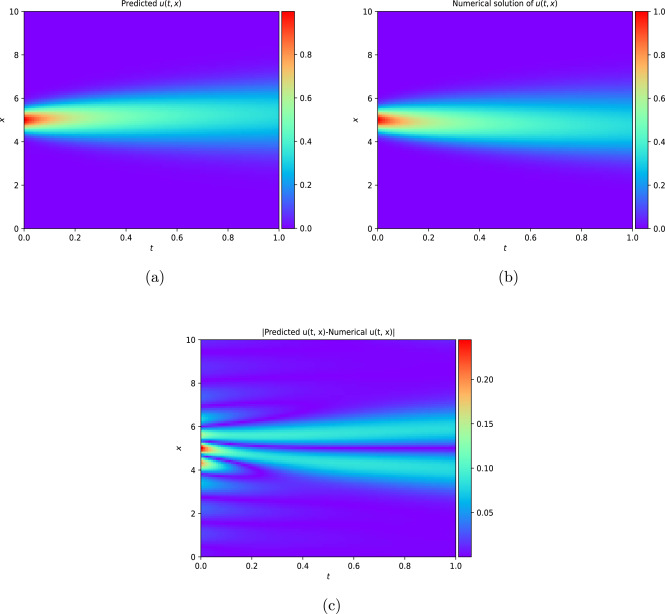
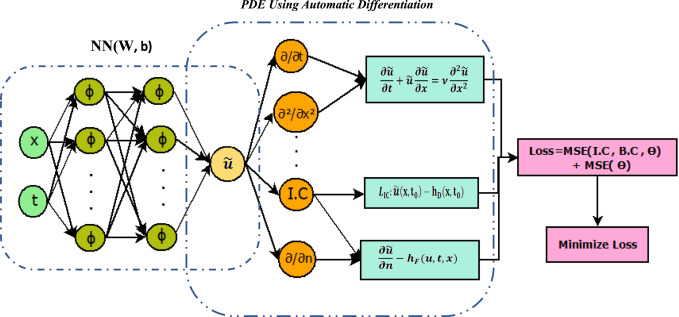
This work introduces a Python implementation of solution of non-liner PDEs arising in the process of arterial blood flow using DeepXDE. The proposed deep learning approach analyzes perturbations in arterial blood flow, with a focus on pressure and radius variations. The research develops a mathematical model for simulating viscoelastic arterial flow, incorporating long wavelength and large Reynolds number assumptions. Leveraging the reductive perturbation method, the study derives nonlinear evolutionary equations for medium resistance, elastic properties, and wall viscosity. By employing state-of-the-art physics-informed deep neural networks, trained via automatic differentiation, the implementation efficiently solves these equations. Bayesian Hyperparameter Optimization identifies the optimal neural network architecture, providing an efficient and accurate alternative to numerical methods for medical machine learning applications.
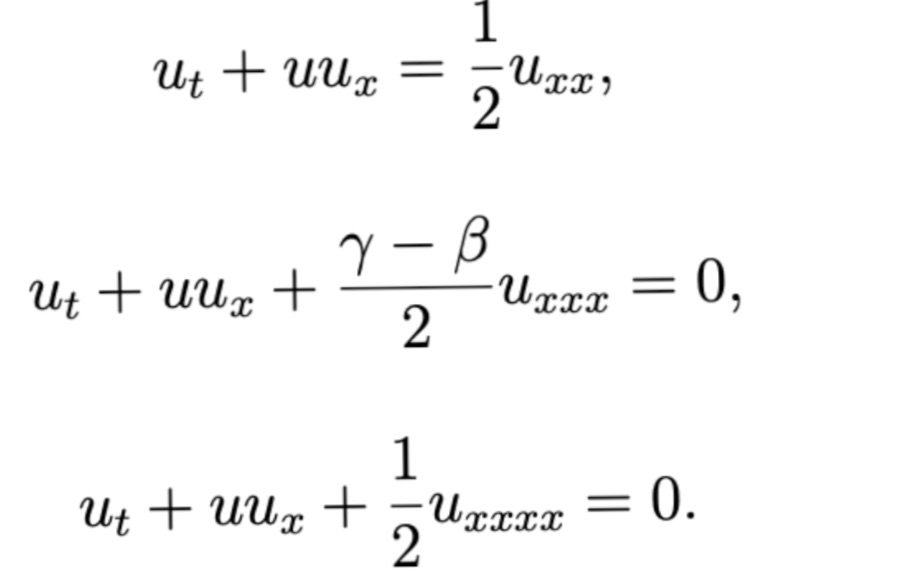
Results
Here are some images showcasing the results: Image source: published Paper link
Project 4: Fully Automated Tumor Segmentation for Brain MRI data using Multiplanner UNet
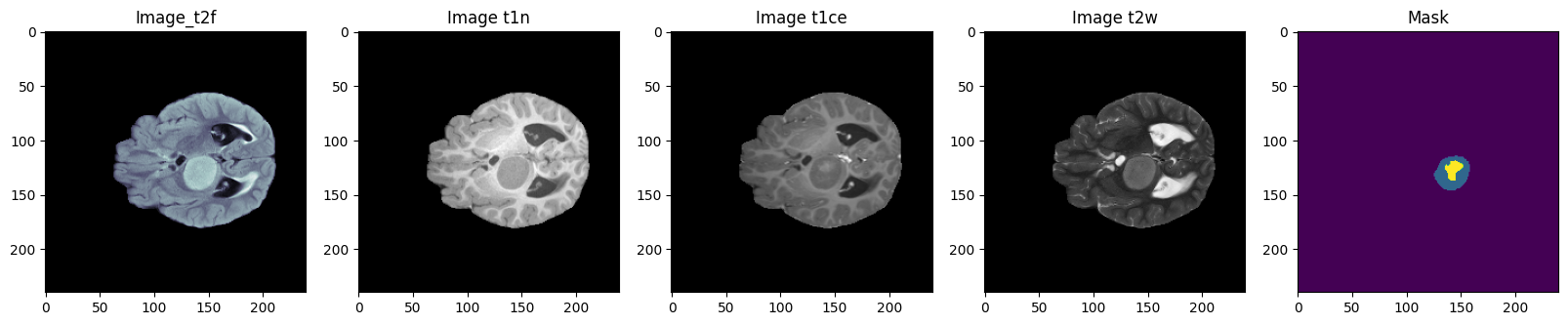
Automated segmentation of distinct tumor regions is critical for accurate diagnosis and treatment planning in pediatric brain tumors. This study evaluates the efficacy of the Multi-Planner U-Net (MPUnet) approach in segmenting different tumor subregions across three challenging datasets: Pediatrics Tumor Challenge (PED), Brain Metastasis Challenge (MET), and Sub-Sahara-Africa Adult Glioma (SSA). These datasets represent diverse scenarios and anatomical variations, making them suitable for assessing the robustness and generalization capabilities of the MPUnet model. By utilizing multi-planar information, the MPUnet architecture aims to enhance segmentation accuracy. Our results show varying performance levels across the evaluated challenges, with the tumor core (TC) class demonstrating relatively higher segmentation accuracy. However, variability is observed in the segmentation of other classes, such as the edema and enhancing tumor (ET) regions. These findings emphasize the complexity of brain tumor segmentation and highlight the potential for further refinement of the MPUnet approach and inclusion of MRI more data and preprocessing.